Sequencing Difficult DNA Templates
|
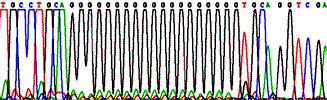
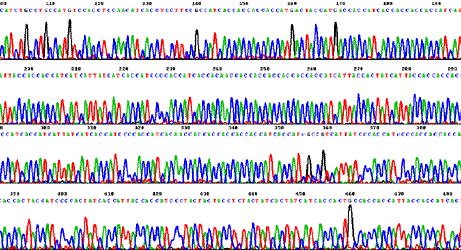
Standard shotgun protocols often fail to produce satisfactory reads with so-called "difficult" templates, such as GC- and AT-rich samples, long simple mono- and di-nucleotide repeats, multiple inverted (hairpin) repeats, and regions with strong stop sites. Our sequencing technology solves many problems with difficult templates using modified protocols with ThermoFidelase and Fimers. Repeats
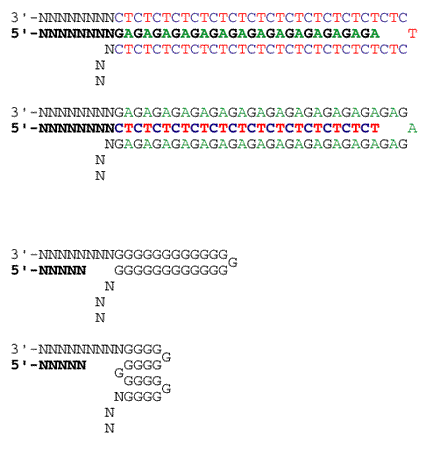
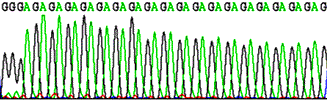
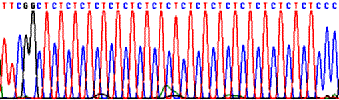
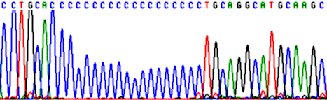
Sequencing through repeated and structured regions: simple nucleotide repeats, Pu and Py repeats.
Sequencing AT-rich templates: plasmids with cloned DNA of Plasmodium falciparum.
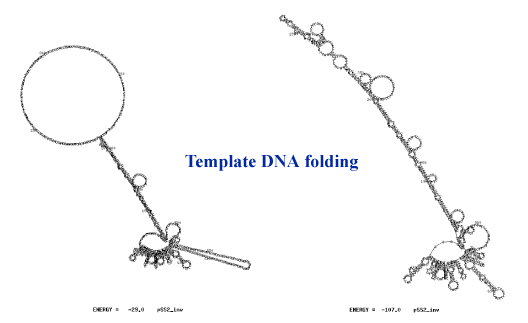
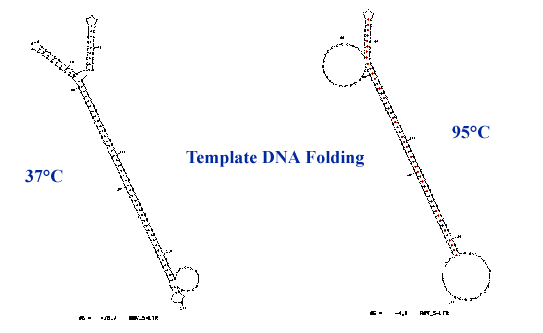
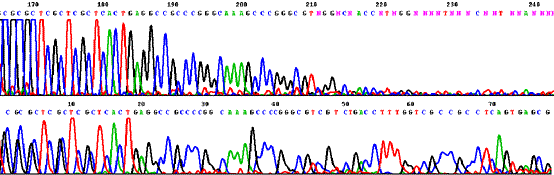
Sequencing through structured regions: long GC rich hairpins (40 kb template)
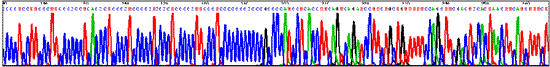
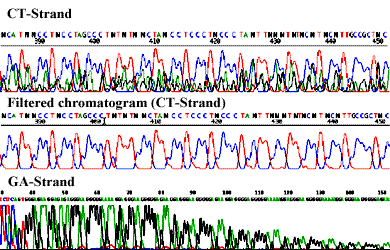
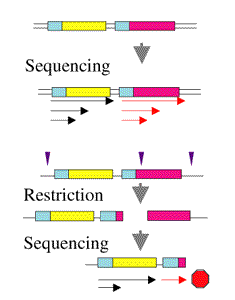
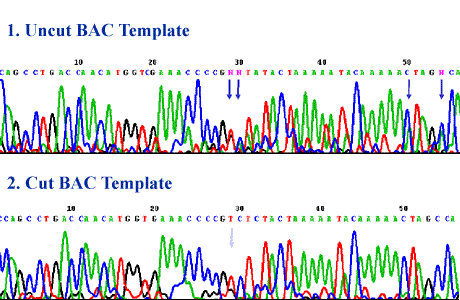
Sequencing through structured regions: Selective sequencing of BAC repeats: getting single reads with multiple priming after treatment of the template DNA with a restrictase. BAC with cloned DNA from Arabidopsis thaliana.
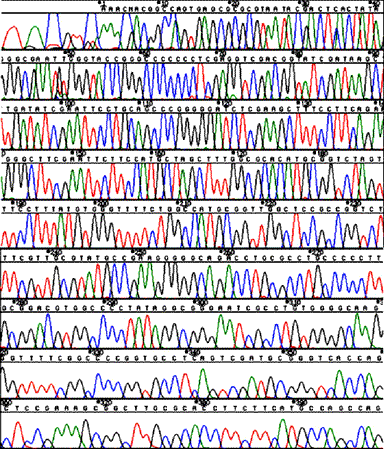
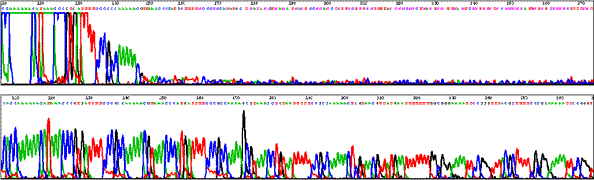
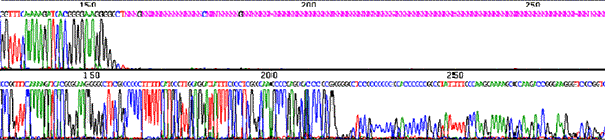
Stops in human DNA clones in BAC.
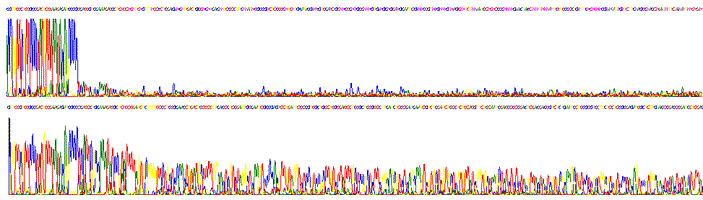
Sequencing through multiple strong stops (40 kb template DNA from Rhodobacter capsulatus).
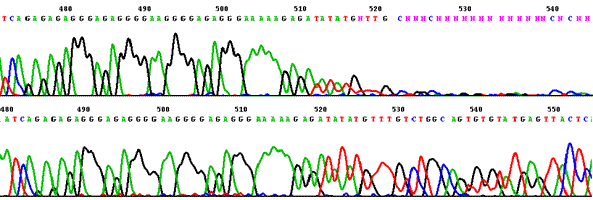
Sequencing through strong stop (template DNA from Bifidobacterium cloned into pUC19).
|